GeomBD
Introduction
A Brownian dynamics (BD) software package that utilizes BD to model molecular diffusion under various experimental environments and biomolecular arrangements.
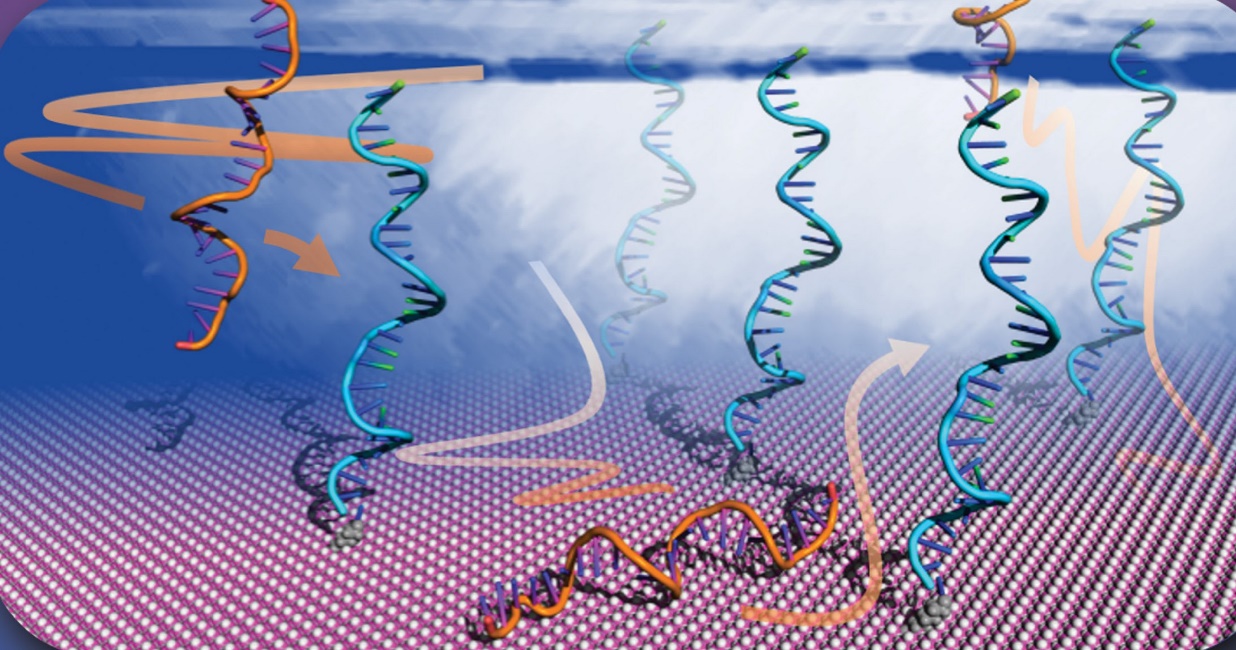
GeomBD3 (GeomBD version 3) is a Brownian dynamics (BD) software package that utilizes BD to model bioengineering and biochemical systems with various experimental environments and biomolecular arrangements. It can be applied to investigate systems up to hundreds of nanometers in scale, such as, enzyme bioconjugates, DNA biosensors, multi-enzyme complexes, and ligand-receptor binding under continuous flow for biocatalysis or using surface plasmon resonance (SPR) assays. The simulations use all-atom, rigid molecular models that diffuse according to overdamped Langevin dynamics and interact through electrostatic with provided salt concentrations, Lennard-Jones, and ligand desolvation potentials. GeomBD3 can consider multiple numbers of user-defined association criteria and calculate molecular association rates, binding residence times, and the statistical and error analysis. Additional tools are provided for the further examinations and visualization, for example, outputting molecular association pathways, diffusion coefficients, intermolecular interaction energies, and intermolecular contact probability maps.
Computational Tools
GeomBD can be performed using the following software tools: