Estimation of relative residence times of protein-ligand complexes using τ-Random Acceleration Molecular Dynamics (τRAMD), implementation in Gromacs
Summary
This tutorial guides the user through the process of setting up and running RAMD simulations for estimation of the relative residence time (τ) of a protein-small molecule complex using the GROMACS software. The procedure is demonstrated for a complex of a low molecular weight compound with the N-terminal domain of the heat shock protein, HSP90.
Important note: This tutorial is for running calculations with Gromacs-RAMD version 1.0. Gromacs-RAMD version 1.0 has a issue with the OpenMP/MPI implementation (see https://github.com/HITS-MCM/gromacs-ramd/issues/11 ); the GPU implementation is not affected.
Gromacs-RAMD version 1.1 (https://github.com/HITS-MCM/gromacs-ramd/releases/tag/gromacs-2020.3-ramd-1.1-rc1) produces output in a different format, requiring small changes to the processing script. It runs with on OpenMP/MPI or GPU.
Gromacs-RAMD version 2.0 (https://github.com/HITS-MCM/gromacs-ramd/releases/tag/gromacs-2020.5-ramd-2.0) requires a somewhat different input format, see here for example input file corresponding to the tutorial. It runs with on OpenMP/MPI or GPU.
Video of a lecture/tutorial, illustrating the main idea, technical aspects, and applications of the tauRAMD approach: https://www.youtube.com/watch?v=kCUyQtoo4cE&feature=youtu.be [2020 MolSSI School on Open Source Software in Rare Event Path Sampling Strategies, July 2020]
Theoretical Background
The use of a high resolution crystal structure of the protein-ligand complex as the starting structure usually gives the most reliable and accurate model of the bound state, which is important for accurate simulation of the bound state. The application of the method to protein-ligand complexes obtained by docking or modeling the ligand binding mode by superposition is possible, but this usually requires longer equilibration times and more extensive sampling of binding poses.
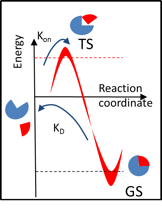
The residence time (\(\tau = 1 / k_\textrm{off}\)) of a complex is defined by the transition barrier energy (\(k_\textrm{off} \sim \exp \left(-\Delta G^\# \right)\)) and therefore depends on the characteristics of the bound (ground state - GS) as well as the transition (TS) states. Thus, simulation of the residence time requires sampling over the bound state (that defines affinity) and dissociation pathways.
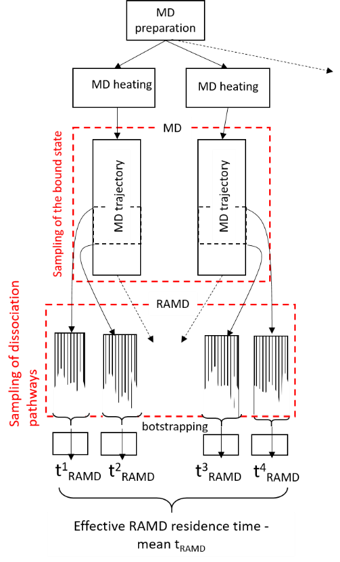
τRAMD provides an efficient simulation procedure for this purpose which is illustrated below. Specifically, several equilibration trajectories provide starting points for simulation of RAMD dissociation trajectories, while RAMD simulation of multiple dissociation trajectories from each starting point provides sampling of possible transition states.
Required Software and Data
Software
- Amber 14 tools and Sander for MD simulations
- GROMACS-RAMD 1.0 v11 ; can be downloaded from: GIT repository
- Python 3.x (libraries: numpy.py; matplotlib, pylab, seaborn, scipy, ParmEd)
- Pymol (or another visualization tool for ligand protonation
- VMD
Scripts and Input files
- Tutorial Instructions in PDF format
- You can download all the input files and scripts required to run this tutorial:
- for GROMACS-RAMD 2.0 get .tar file here.
- for GROMACS-RAMD 1.1 get .tar file here.
- the latest version of the script for computing residence time can be also updated from here tauRAMD.py
Type the following command to extract RAMD_tutorial_files-Gromacs.tar in a directory called /home/ramd/
tar -xvf RAMD_tutorial_files-Gromacs.tar -C /home/ramd/
Organization of the file system for τRAMD simulations
The directories are organized in this tutorial as follows :
- AMBER/ #contain topology and coordinate files
- Gromacs/
- Replica1/ #contain conventional MD equilibration trajectories
Replica2/
Replica3/
.....
- Replica1/ #contain conventional MD equilibration trajectories
- RAMD/
- Replica1
- TRJ1-1/ #contain conventional MD equilibration trajectories
TRJ-2/
TRJ-3/
....
- TRJ1-1/ #contain conventional MD equilibration trajectories
TRJ-2/
TRJ-3/
....
- TRJ1-1/ #contain conventional MD equilibration trajectories
- Replica1
Description of the scripts and input files used during this tutorial
Preparation of the protein and ligand structures
complex.pdb – starting coordinates of the protein-ligand complex in PDB format
Generation of the topology and coordinate files for protein and ligand
tleap_ligand_in - tleap input file for generation of the ligand parameters
tleap_all_in - tleap input file for generation of the topology and coordinate files of the complex
Energy minimization, heating and short equilibration with restrains using AMBER
amber_prep.sh
Heating and equilibration using Gromacs (first using Berendsen thermostat, then using Nose-Hoover)
gromacs.mdp
gromacs1.mdp
Simulation of dissociation using RAMD
RAMD-force.sh
Analysis of trajectories
tauRAMD.py
Procedure
Step 1: Preparation of the protein and ligand structures
Partition the PDB format file (complex.pdb in our case) of the coordinates of the protein-ligand complex (without hydrogens added) into two files: protein (without hydrogens) and ligand (inhibitor is named as INH in complex.pdb file) using the UNIX grep command.
grep ^ATOM complex.pdb > protein.pdb
grep INH complex.pdb > ligand.pdb
Protonate the ligand using pymol and save it as ligand_H.pdb; then remove all connectivity lines in the file.
- For protonation, load the ligand.pdb file into Pymol and click Action -> hydrogens -> add to add hydrogen atoms. Then save the molecule again as ligand.pdb (File -> Save molecule -> ligand -> OK). Instead of Pymol, another software (such as Chimera, Maestro Schrödinger, MOE) can be used. However, assignment of the protonation state is not always unambiguous and generally requires computation of pKa values. In the present case, the Pymol procedure is sufficient.
Select crystallographic water molecules within 5 Angstroms of the ligand, protonate them, and save them in the file water.pdb
- To select crystallographic water molecules, load the file complex.pdb into Pymol and type the following command in the Pymol command line utility: select resname HOH within 5 of resname INH
- This will create a selection named sele with water molecules within 5 Angstroms of the ligand. Then add hydrogen atoms to the selection (Action -> hydrogen -> add) and save the sele as water.pdb (File -> Save molecule -> sele -> OK)
Rename hydrogen atoms in the file water.pdb to make the atom naming scheme the same as for the protein (as the schemes are different in Amber and pymol):
sed "s/H01/H1 /" water.pdb > water1.pdb
sed "s/H02/H2 /" water1.pdb > water2.pdb
Step 2: Generation of protein and ligand parameters
Use the Amber tool antechamber to generate a mol2 format file from the pdb file of the ligand
antechamber -i ligand_H.pdb -fi pdb -o ligand_H.mol2 -fo mol2 -c bcc -s 2 -nc -1
- Antechamber generates a mol2 file that contains partial charges computed using the semiempirical BCC method (see http://ambermd.org/antechamber/ac.html for more details). The partial charges of the ligand can be also computed using the RESP web server (http://upjv.q4md-forcefieldtools.org/REDServer-Development/ ). Since the coordinates of the output mol2 file from the RESP web-server will differ from the original, this file cannot be used directly for simulations. Instead, the RESP partial charges from the mol2 file should be used to replace the partial charges of the ligand_H.mol2 file.
Use parmchk from Amber tools to determine the bond and atom types of the ligand (file INH.frcmod):
parmchk2 -i ligand_H.mol2 -f mol2 -o INH.frcmod
Use tleap from Amber tools to generate the ligand topology file, coordinate and pdb files (INH.lib, INH.crd, INH.pdb):
tleap -f tleap_ligand_in
Combine the protein, ligand, and crystallographic water PDB files into one file in which their coordinates are separated by TER lines:
grep ATOM protein.pdb > prot_lig.pdb
echo "TER" >> prot_lig.pdb
grep INH INH.pdb >> prot_lig.pdb
echo "TER" >> prot_lig.pdb
grep HOH water2.pdb >> prot_lig.pdb
Solvate the system and add ions using tleap:
Number of added ions depends on protonation states of protein and ligand (different methods may give different protonation states). Ions should be added to make the system electroneutral (system charge should be 0) and to make the system's salt concentration equal to 0.150 M. With this in mind, number of added ions in tleap_all_in file should be changed accordingly.
Please consult eg: calculate number of ions on ambermd.org or heretleap -f tleap_all_in
which outputs topology (ref.prmtop), PDB (ref.pdb) and coordinate (ref.impcrd) files in Amber format.
(files above depend on protonation method chosen for ligand and protein)Step 3: Energy minimization, heating, and equilibration of the protein-ligand complex using Amber
The energy minimization procedure consists of four steps: first with restraints on all protein and ligand heavy atoms (with force constants of 500, 50, and 5 kcal/mol A2) and then without restraints.
The system is gradually heated to 300K keeping heavy atoms restrained. Finally, the system is equilibrated in four short equilibration runs: with restraind decreasing from 50 kcal/mol A2 to 10 kcal/mol A2 and then 2 kcal/mol A2, and without restrains"
The Input files are prepared using the amber_prep.sh script and minimization is performed using the Sander module of Amber. Run the amber_prep.sh script through the command line as:
sh amber_prep.sh
The output coordinate file ref-equil-NPT.crd will be used as an input for the next heating/equilibration step and in RAMD simulations.
Step 4: Heating and equilibration of the protein-ligand complex using Gromacs
For sampling of the bound state in RAMD, multiple starting points (i.e. Gromacs *cpp restart files) usually need to be prepared.
- There are two strategies – either repeating the heating and equilibration steps several times (due to the random distribution in velocity assignment, each trajectory will be slightly different), or breaking the long equilibration run into parts and using intermediate states. These strategies can be combined to get better sampling. However, in the present example, we will prepare just one RAMD starting point for simplicity.
The Amber topology file and the output restart file need to be transferred into Gromacs-format files as follows:
- Generate the AMBER restart file for each simulated system:
You will need AMBER topology (*.prmtop) and input restart (*.rst7) files (restart file *rst7 can be obtained from a coordinate file *crd using cpptraj ) generated after energy minimization, heating, and a short equilibration (see, for example, the procedure described in the previous section) .
Here is an example of the input file for cpptraj to transfer *crd file to *rst7:parm ref.prmtop
trajin ref-equil-NPT.crd
trajout ref-equil.rst7
run
- Generate the Gromacs topology and coordinate files:
Launch Python and use the ParmEd library (http://parmed.github.io/ParmEd/html/index.html) to prepare Gromacs topology (*.top) and *.gro files as follows:import parmed as pmd
amber = pmd.load_file('ref.prmtop', 'ref-equil.rst7')
amber.save('gromacs.top')
amber.save('gromacs.gro')
- Generate the Gromacs index file:
First, the index.ndx file with appropriate subsystems (called “groups” in Gromacs) needs to be generated using the interactive programe make_ndx of Gromacs:gmx make_ndx -f gromacs.gro -o index.ndx
Subsystems can be: all membrane lipids, ‘Protein_ligand’, and ‘Water_and_ions’ . For example, for the HSP90-inhibitor (INH) system, the subsystems are ‘Protein_INH’ and ‘Water_and _ions’
Perform the first equilibration step (using the Berendsen thermostat) The Gromacs execution command may depend on the system environment and options used for compilation. The simplest command is:
gmx grompp -f gromacs.mdp -c gromacs.gro -o gromacs0.tpr -n index.ndx -p gromacs.top -maxwarn 5 -n index.ndx
gmx mdrun -s gromacs0.tpr -ntmpi 1 -ntomp16 -maxh 24
Perform the second equilibration step (using the Nose-Hoover thermostat and the Parrinello-Rahman barostat) to generate starting replicas. This step can be repeated to generate multiple replicas:
gmx grompp -f gromacs1.mdp -t state.cpt -c gromacs0.tpr -o gromacs1.tpr -n index.ndx -p gromacs.top -maxwarn 5;
gmx mdrun -s gromacs1.tpr -ntmpi 1 -ntomp 16 -maxh 24
Note that Equilibration replicas should be generated in the directories: Replica1, Replica2, etc. (see Section Organization of the file system for tauRAMD simulations) in this step, with the output coordinates from the first equilibration step as input and using a Maxwell distribution of velocities by specifying the parameter “gen_vel = yes” for each replica (I.e. to start new trajectories).
Also note that if you are using a membrane system the 'pcoupltype' in gromacs1.mdp must be changed to 'semiisotropic'. The same parameter has to be changed in gromacs_ramd.mdp for the following step.
Step 5: Generation of the ligand dissociation trajectories using Gromacs-RAMD
You will use an input file gromacs_ramd.mdp that is adjusted to the current HSP90 case
For any other system, one has to change the following parameters in the input file:
ramd-force ; force to be applied - in kJ mol-1 nm-1 ;
ramd-ligand ; ligand residue name;
ramd-max-dist ; maximal distance of ligand COM displacement when RAMD simulations are stopped /samp>
Use the script Submit_15_jobs_gromacs-ramd.sh to set up the simulations and store them in the respective RAMD replica directories together with the gromacs_ramd.mdp file
Note that this script assumes that the file structure is as given in above in the Section "Organization of the file system for τRAMD simulations".
The job_gromacs-ramd.sh script automatically generates directories for the simulated trajectories and submits 15 trajectories for simulation.The lines marked “#to be edited<---” must be edited for the particular job submission environment used
Run RAMD simulations trajectory as follows:
./Submit_15_jobs_gromacs-ramd.sh/samp>
Step 6: Statistical analysis of the ligand dissociation trajectories
Extract the time when the ligand -protein distance reaches the maximum value (defined as 40 Angstroms in the RAMD input file) from each trajectory using the following command:
grep "GROMACS will be stopped after" TRJ*/md.log > times_1.dat
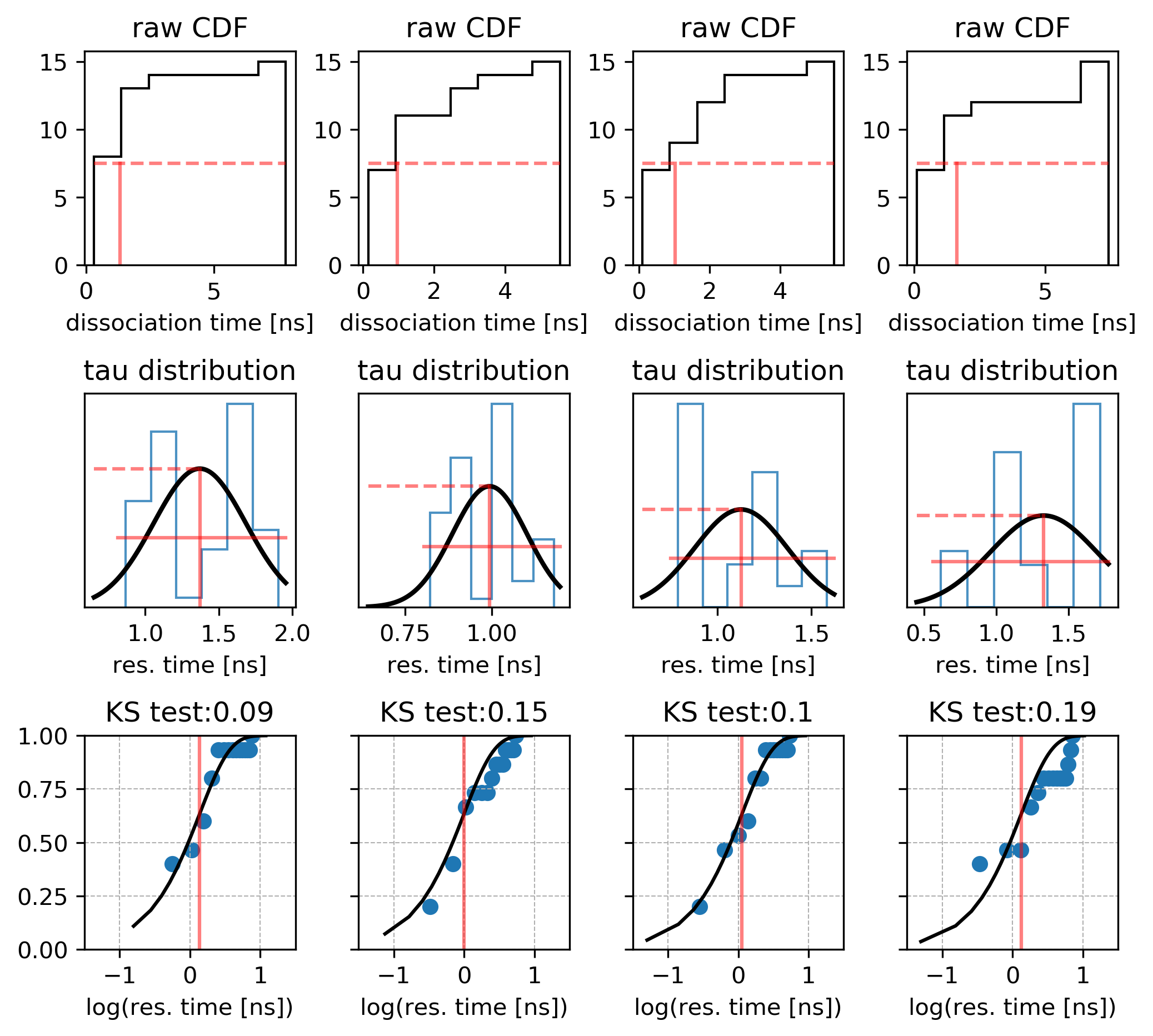
Plot the distribution of these times given in the times_1.dat file as a histogram. Ideally, the distribution should converge to an exponentially decreasing curve if there is only one dissociation route. The effective residence time is defined as the time when 50% of the trajectories have dissociated.
Use the bootstrapping method to derive the relative dissociation time from the trajectories generated by RAMD by running Python script using the following command:
python tauRAMD.py times_1.dat
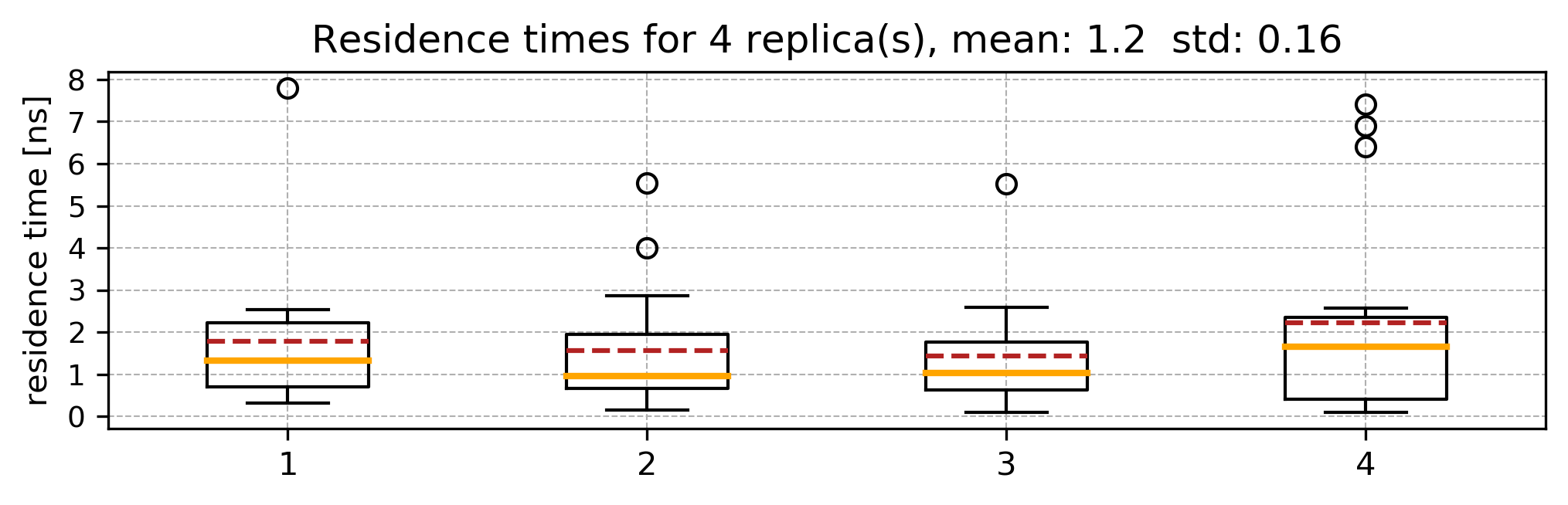
The mean and standard deviation values printed in the standard output correspond to the relative residence time in ns and the standard deviation.
Running several sets of RAMD simulations starting from different equilibration points will provide a set of the residence time distributions, like this: