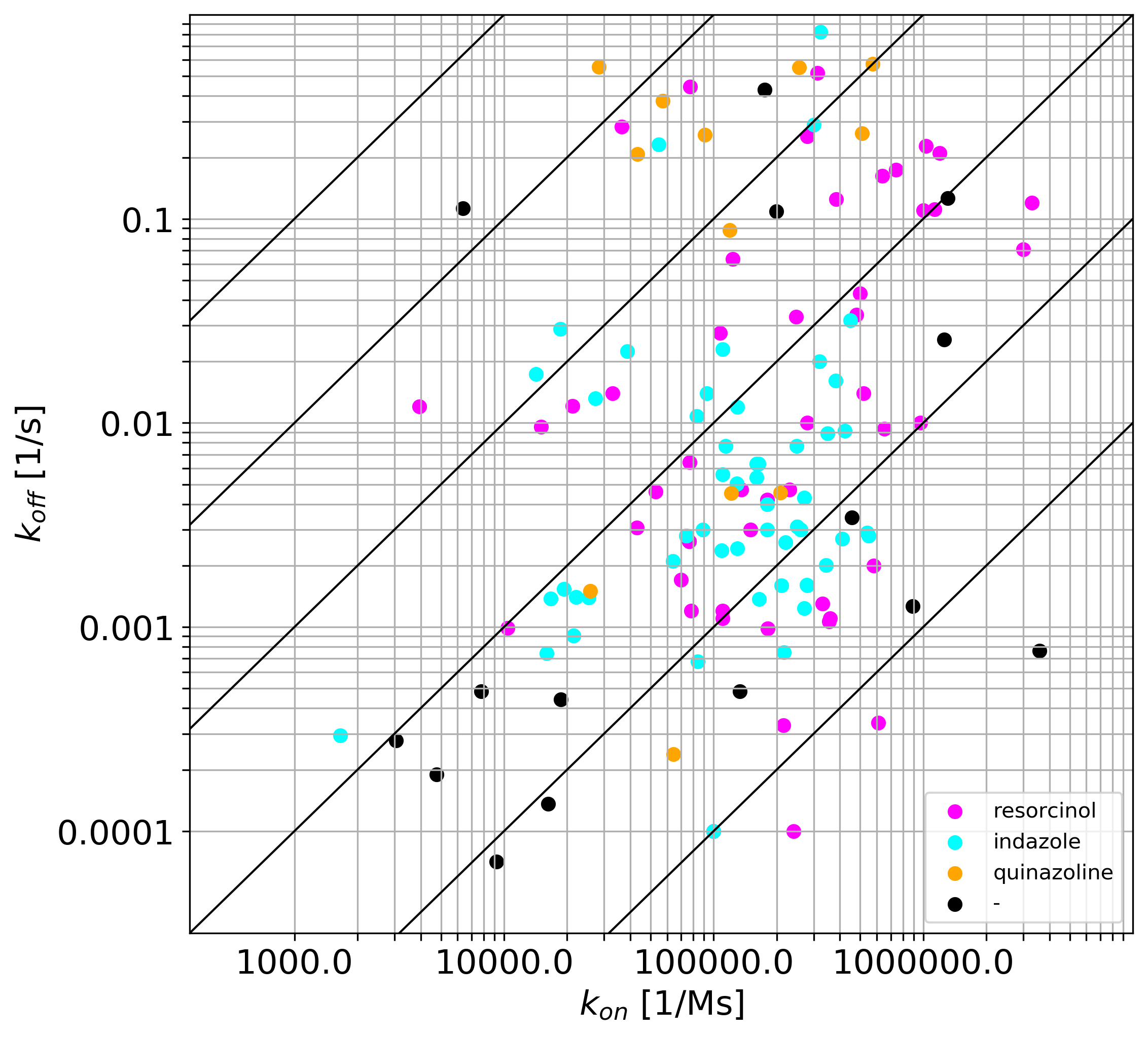
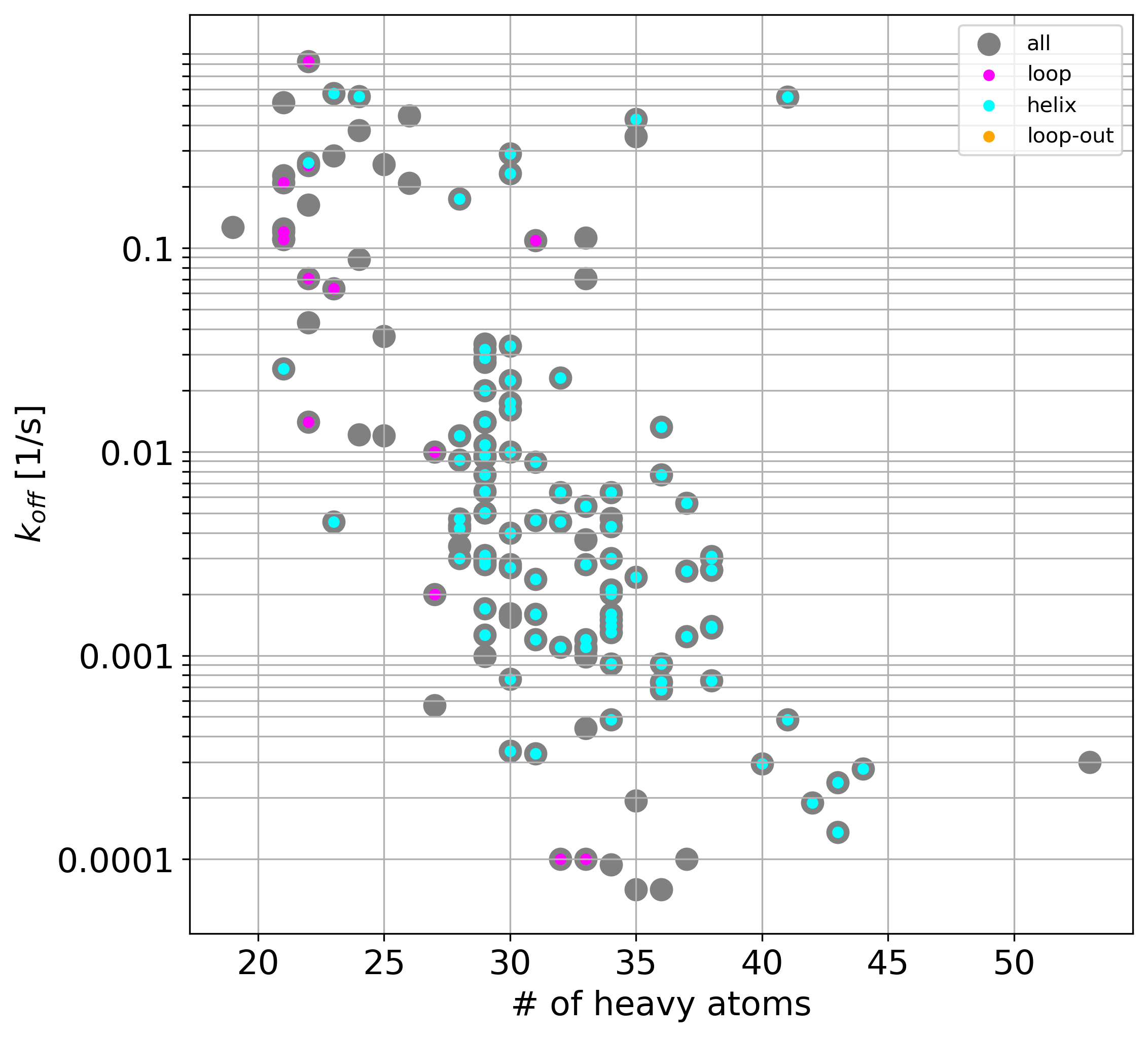
HSP90 - SPR
Introduction
Please download data file here: HSP90 - SPR
(uploaded on 2020-01-30).
Binding kinetics data set for HSP90 inhibitors
Author: Daria Kokh (daria.kokh@h-its.org)
The Excel file contains a set of kinetic rate constants and SMILE strings for more than 140 small molecule inhibitors of the N-terminal domain of heat shock protein 90 (HSP90).
Date were obtained by SPR measurements (using a single protocol) as a part of the K4DD project and were reported in the four publications listed below.
 |
 |
 |
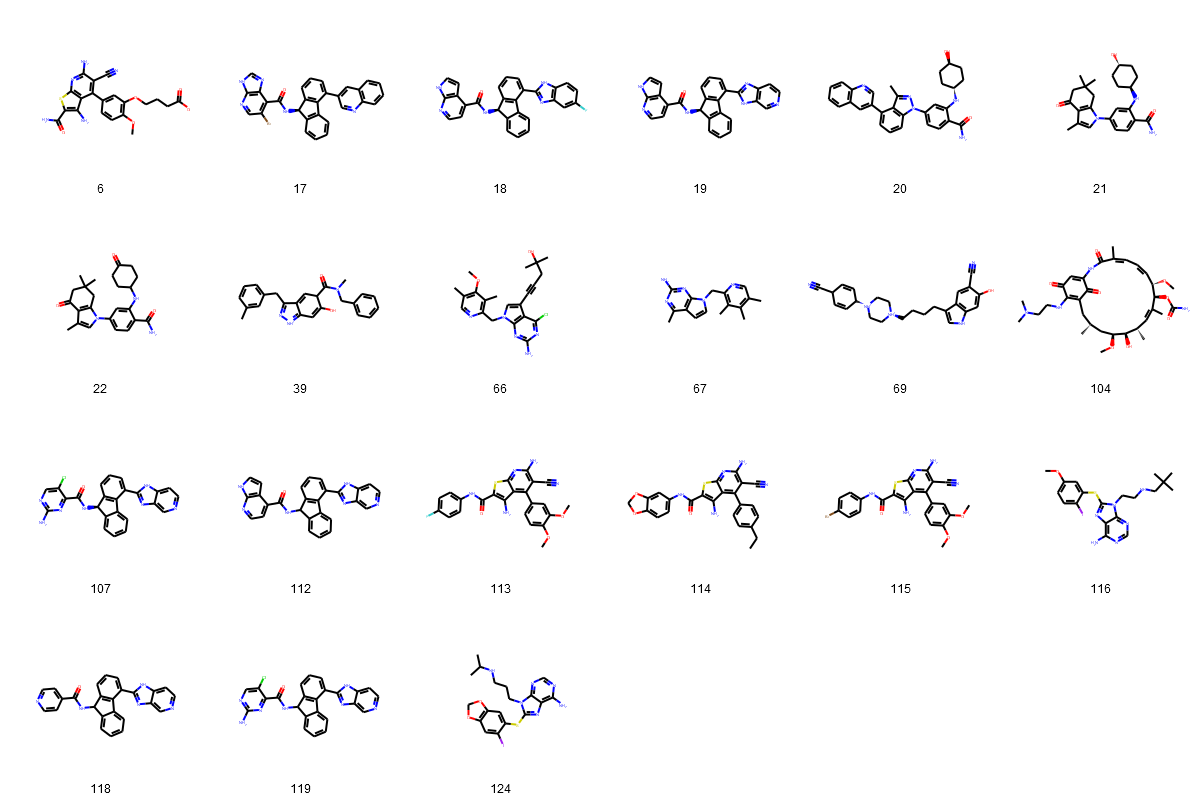 |
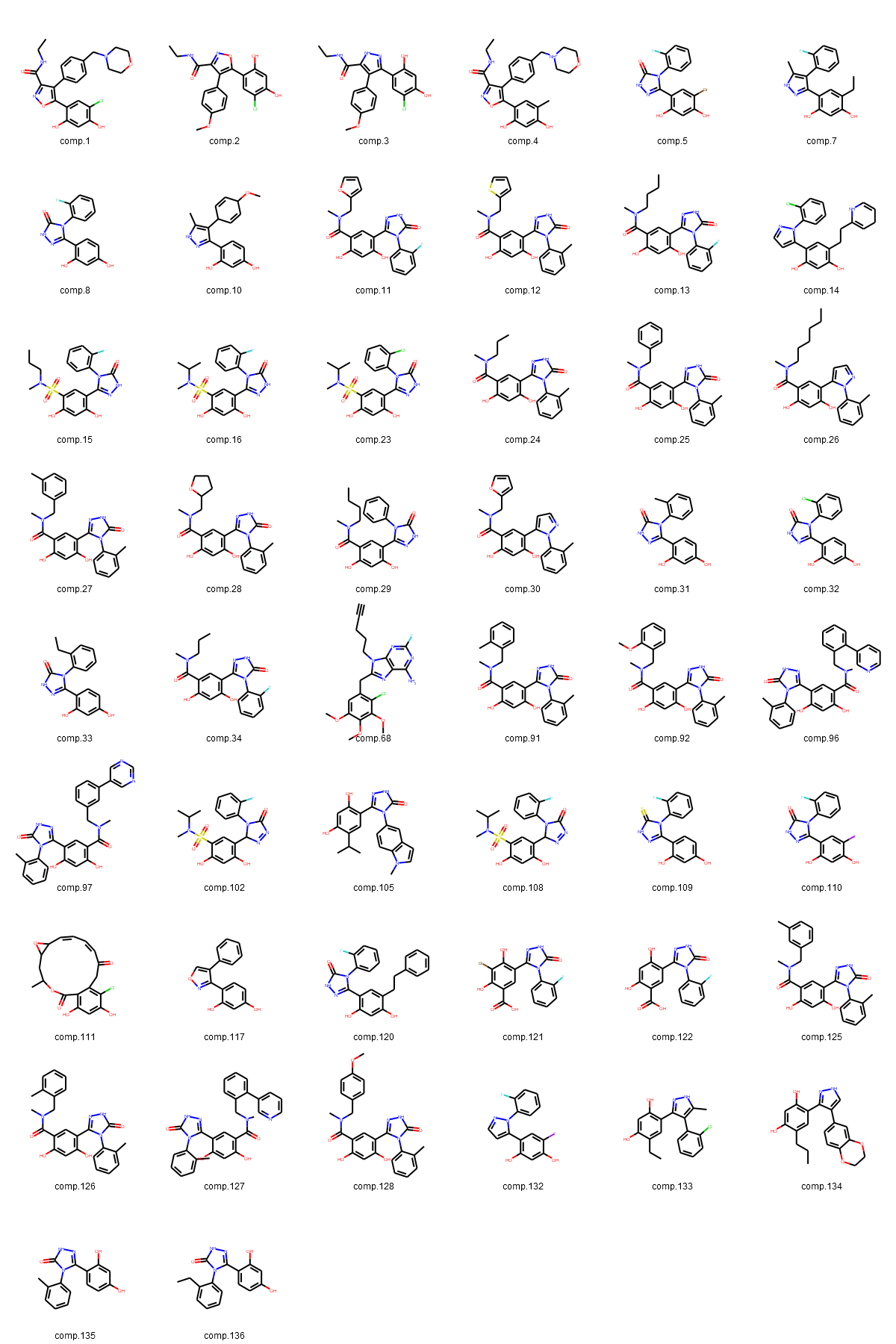
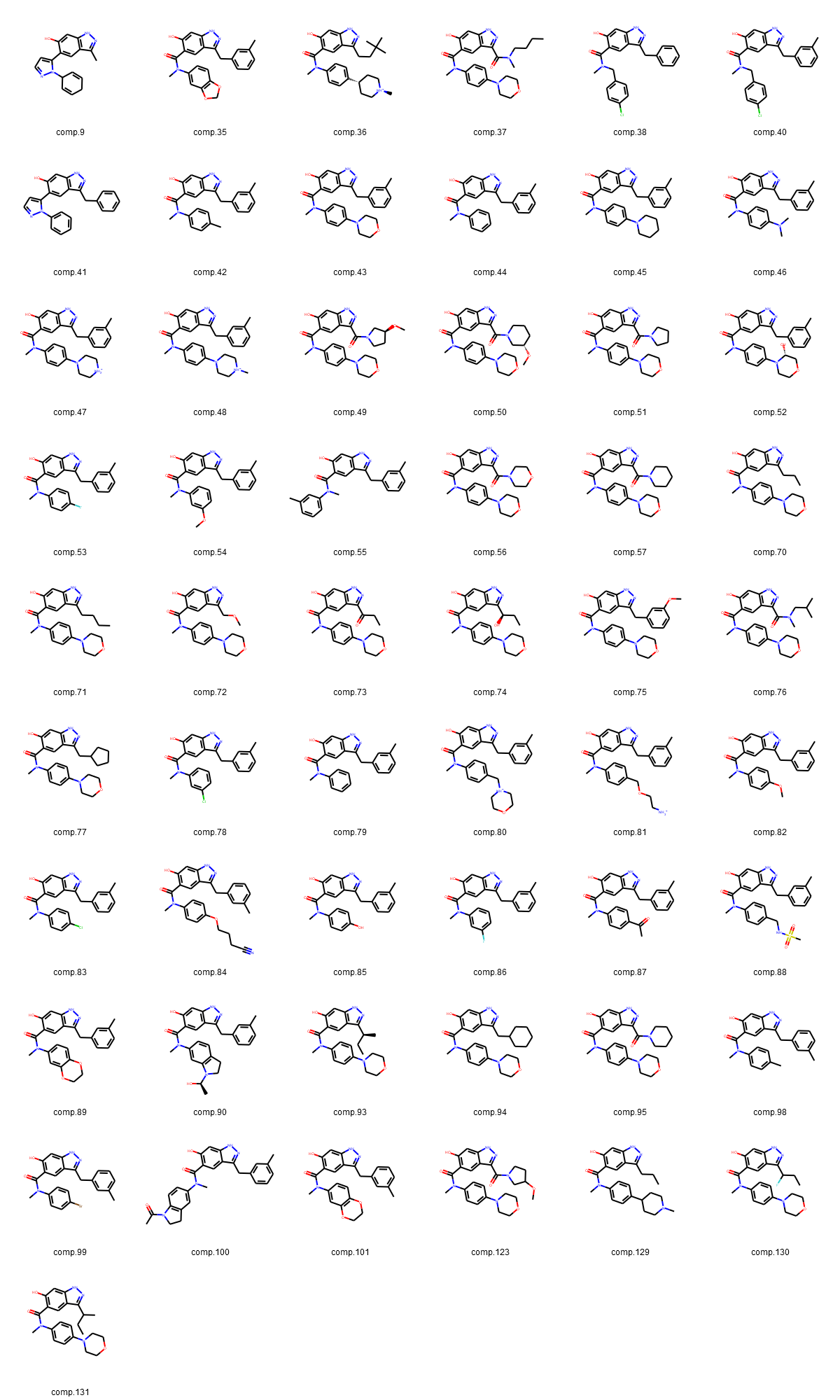
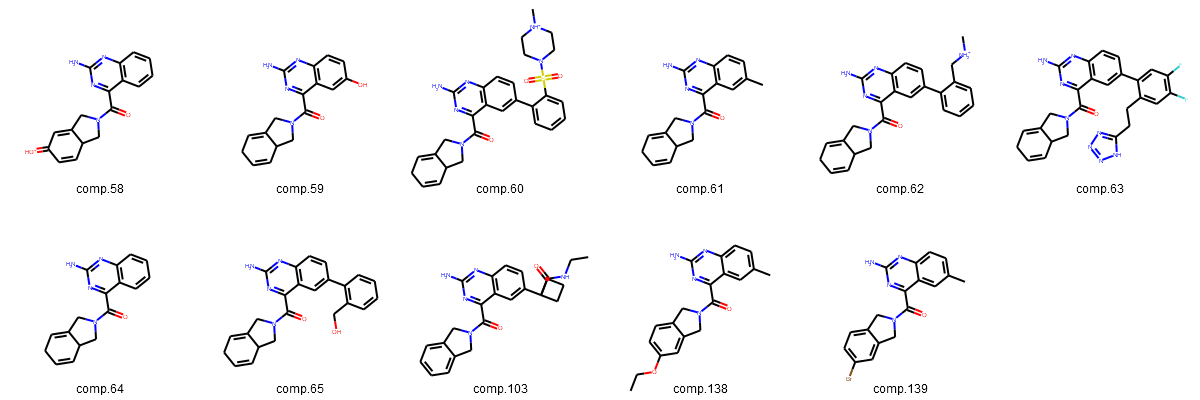
Fig. 3: (A-D): 2D images of compounds (separated into three largest groups by the core fragments: resorcinol, indazole, quinazoline and one group of compounds with various cores).
Data related to this data set:
- Chao Xu, Xianglei Zhang, Lianghao Zhao, Gennady M. Verkhivker, and Fang Bai: Accurate Characterization of Binding Kinetics and Allosteric Mechanisms for the HSP90 Chaperone Inhibitors Using AI-Augmented Integrative Biophysical Studies. JACS Au 2024, XX:XXXX-XXXX.
Data were collected from the following publications:
- Kokh DB, Amaral M, Bomke J, Grädler U, Musil D, Buchstaller HP, Dreyer MK, Frech M, Lowinski M, Vallee F, et al.: Estimation of Drug-Target Residence Times by τ-Random Acceleration Molecular Dynamics Simulations. J Chem Theory Comput 2018, 14:3859–3869. (see method description 'τ-RAMD' in kbbox)
- Kokh Daria B., Kaufmann Tom, Kister Bastian, Wade Rebecca C., Machine Learning Analysis of τRAMD Trajectories to Decipher Molecular Determinants of Drug-Target Residence Times, Front. Mol. Biosci. (2019) 6,1-17 (see method description 'τ-RAMD' in kbbox)
- Schuetz, D. A.; Richter, L.; Amaral, M.; Grandits, M.; Grädler, U.; Musil, D.; Buchstaller, H.-P.; Eggenweiler, H.-M.; Frech, M.; Ecker, G. F. Ligand Desolvation Steers on-Rate and Impacts Drug Residence Time of Heat Shock Protein 90 (Hsp90) Inhibitors. J. Med. Chem. 2018, 61, 4397–4411. (see method description 'steered molecular dynamics' in kbbox)
- Schuetz DA, Bernetti M, Bertazzo M, Musil D, Eggenweiler HMH-M, Recanatini M, Masetti M, Ecker GF, Cavalli A: Predicting Residence Time and Drug Unbinding Pathway through Scaled Molecular Dynamics. J Chem Inf Model 2019, 59:535–549. (see method description 'smoothed or scaled molecular dynamics' in kbbox)
- Wolf S, Amaral M, Lowinski M, Vallée F, Musil D, Güldenhaupt J, Dreyer MK, Bomke J, Frech M, Schlitter J, et al.: Estimation of Protein-Ligand Unbinding Kinetics Using Non-Equilibrium Targeted Molecular Dynamics Simulations. J Chem Inf Model 2019, 59:5135–5147. (see methods description 'targeted molecular dynamics' in kbbox)
K4DD project was supported by EU/EFPIA Innovative Medicines Initiative (IMI) Joint Undertaking, K4DD (grant no. 115366)